ALIGNN Tutorial#
Install ALIGNN:
pip install alignn
from alignn.ff.ff import AlignnAtomwiseCalculator,default_path
model_path = default_path()
calc = AlignnAtomwiseCalculator(path=model_path)
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
Cell In[1], line 1
----> 1 from alignn.ff.ff import AlignnAtomwiseCalculator,default_path
3 model_path = default_path()
4 calc = AlignnAtomwiseCalculator(path=model_path)
File /media/colin/Shared/colin/git/materials-ml-workshop/env/lib/python3.10/site-packages/alignn/ff/ff.py:34
32 from jarvis.db.jsonutils import loadjson
33 from alignn.graphs import Graph
---> 34 from alignn.models.alignn_atomwise import ALIGNNAtomWise, ALIGNNAtomWiseConfig
35 from jarvis.analysis.defects.vacancy import Vacancy
36 import numpy as np
File /media/colin/Shared/colin/git/materials-ml-workshop/env/lib/python3.10/site-packages/alignn/models/alignn_atomwise.py:14
11 import torch
13 # from dgl.nn.functional import edge_softmax
---> 14 from pydantic.typing import Literal
15 from torch import nn
16 from torch.nn import functional as F
ImportError: cannot import name 'Literal' from 'pydantic.typing' (/media/colin/Shared/colin/git/materials-ml-workshop/env/lib/python3.10/site-packages/pydantic/typing.py)
from ase import Atom, Atoms
import numpy as np
import matplotlib.pyplot as plt
lattice_params = np.linspace(3.5, 3.8)
def make_fcc_copper(a=3.6):
cu_atoms = Atoms([Atom('Cu', (0, 0, 0))],
cell=0.5 * a * \
np.array([[1.0, 1.0, 0.0],
[0.0, 1.0, 1.0],
[1.0, 0.0, 1.0]]),
pbc=True)
return cu_atoms
fcc_energies = []
for a in lattice_params:
atoms = make_fcc_copper(a)
atoms.set_tags(np.ones(len(atoms)))
atoms.calc = calc
e = atoms.get_potential_energy()
fcc_energies.append(e)
import matplotlib.pyplot as plt
%matplotlib inline
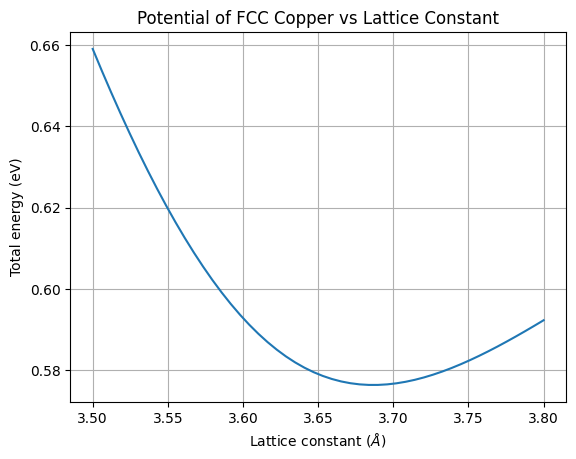
plt.title('Potential of FCC Copper vs Lattice Constant')
plt.grid()
plt.plot(lattice_params, fcc_energies)
plt.xlabel('Lattice constant ($\AA$)')
plt.ylabel('Total energy (eV)')
plt.show()
from mp_api.client import MPRester
from pymatgen.io.ase import AseAtomsAdaptor as aaa
MPID = 'mp-20674' # Materials Project ID number for YBCO-123
with MPRester() as mpr:
structure = mpr.get_structure_by_material_id(MPID)
raw_atoms = aaa.get_atoms(structure) # convert pymatgen to ase
from ase.visualize import view
view(raw_atoms, viewer='x3d')
from alignn.ff.ff import AlignnAtomwiseCalculator, default_path
from ase.optimize import BFGS, BFGSLineSearch
from ase.io import read
import os
def alignn_ff_relax(raw_atoms, nsteps=5,
filename='alignn_ff',
workdir='.'):
atoms = raw_atoms.copy()
atoms.set_tags(np.ones(len(atoms)))
relax_calc = AlignnAtomwiseCalculator(path=default_path())
atoms.calc = relax_calc
init_energy = atoms.get_potential_energy()
dyn = BFGS(atoms,
trajectory=os.path.join(workdir,f'{filename}.traj'),
restart=os.path.join(workdir, f'{filename}.pckl'))
dyn.run(fmax=0.0065, steps=nsteps)
final_energy = atoms.get_potential_energy()
print(f'E_init = {init_energy}')
print(f'E_final = {final_energy}')
return atoms
relaxed_atoms = alignn_ff_relax(raw_atoms)
model_path /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/alignn/ff
Step Time Energy fmax
BFGS: 0 14:30:29 -50.884412 0.0114
BFGS: 1 14:30:30 -50.220586 0.0127
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In [33], line 26
22 print(f'E_final = {final_energy}')
24 return atoms
---> 26 relaxed_atoms = alignn_ff_relax(raw_atoms)
Cell In [33], line 18, in alignn_ff_relax(raw_atoms, nsteps, filename, workdir)
13 init_energy = atoms.get_potential_energy()
15 dyn = BFGS(atoms,
16 trajectory=os.path.join(workdir,f'{filename}.traj'),
17 restart=os.path.join(workdir, f'{filename}.pckl'))
---> 18 dyn.run(fmax=0.0065, steps=nsteps)
19 final_energy = atoms.get_potential_energy()
21 print(f'E_init = {init_energy}')
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/ase/optimize/optimize.py:269, in Optimizer.run(self, fmax, steps)
267 if steps:
268 self.max_steps = steps
--> 269 return Dynamics.run(self)
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/ase/optimize/optimize.py:156, in Dynamics.run(self)
149 def run(self):
150 """Run dynamics algorithm.
151
152 This method will return when the forces on all individual
153 atoms are less than *fmax* or when the number of steps exceeds
154 *steps*."""
--> 156 for converged in Dynamics.irun(self):
157 pass
158 return converged
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/ase/optimize/optimize.py:132, in Dynamics.irun(self)
129 self.call_observers()
131 # run the algorithm until converged or max_steps reached
--> 132 while not self.converged() and self.nsteps < self.max_steps:
133
134 # compute the next step
135 self.step()
136 self.nsteps += 1
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/ase/optimize/optimize.py:274, in Optimizer.converged(self, forces)
272 """Did the optimization converge?"""
273 if forces is None:
--> 274 forces = self.atoms.get_forces()
275 if hasattr(self.atoms, "get_curvature"):
276 return (forces ** 2).sum(
277 axis=1
278 ).max() < self.fmax ** 2 and self.atoms.get_curvature() < 0.0
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/ase/atoms.py:788, in Atoms.get_forces(self, apply_constraint, md)
786 if self._calc is None:
787 raise RuntimeError('Atoms object has no calculator.')
--> 788 forces = self._calc.get_forces(self)
790 if apply_constraint:
791 # We need a special md flag here because for MD we want
792 # to skip real constraints but include special "constraints"
793 # Like Hookean.
794 for constraint in self.constraints:
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/ase/calculators/abc.py:23, in GetPropertiesMixin.get_forces(self, atoms)
22 def get_forces(self, atoms=None):
---> 23 return self.get_property('forces', atoms)
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/ase/calculators/calculator.py:737, in Calculator.get_property(self, name, atoms, allow_calculation)
735 if not allow_calculation:
736 return None
--> 737 self.calculate(atoms, [name], system_changes)
739 if name not in self.results:
740 # For some reason the calculator was not able to do what we want,
741 # and that is OK.
742 raise PropertyNotImplementedError('{} not present in this '
743 'calculation'.format(name))
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/alignn/ff/ff.py:165, in AlignnAtomwiseCalculator.calculate(self, atoms, properties, system_changes)
163 num_atoms = j_atoms.num_atoms
164 g, lg = Graph.atom_dgl_multigraph(j_atoms)
--> 165 result = self.net((g.to(self.device), lg.to(self.device)))
166 # print ('stress',result["stress"].detach().numpy())
167 self.results = {
168 "energy": result["out"].detach().cpu().numpy() * num_atoms,
169 "forces": result["grad"].detach().cpu().numpy(),
(...)
177 "magmoms": np.zeros(len(atoms)),
178 }
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/torch/nn/modules/module.py:1130, in Module._call_impl(self, *input, **kwargs)
1126 # If we don't have any hooks, we want to skip the rest of the logic in
1127 # this function, and just call forward.
1128 if not (self._backward_hooks or self._forward_hooks or self._forward_pre_hooks or _global_backward_hooks
1129 or _global_forward_hooks or _global_forward_pre_hooks):
-> 1130 return forward_call(*input, **kwargs)
1131 # Do not call functions when jit is used
1132 full_backward_hooks, non_full_backward_hooks = [], []
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/alignn/models/alignn_atomwise.py:311, in ALIGNNAtomWise.forward(self, g)
309 # ALIGNN updates: update node, edge, triplet features
310 for alignn_layer in self.alignn_layers:
--> 311 x, y, z = alignn_layer(g, lg, x, y, z)
313 # gated GCN updates: update node, edge features
314 for gcn_layer in self.gcn_layers:
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/torch/nn/modules/module.py:1130, in Module._call_impl(self, *input, **kwargs)
1126 # If we don't have any hooks, we want to skip the rest of the logic in
1127 # this function, and just call forward.
1128 if not (self._backward_hooks or self._forward_hooks or self._forward_pre_hooks or _global_backward_hooks
1129 or _global_forward_hooks or _global_forward_pre_hooks):
-> 1130 return forward_call(*input, **kwargs)
1131 # Do not call functions when jit is used
1132 full_backward_hooks, non_full_backward_hooks = [], []
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/alignn/models/alignn_atomwise.py:171, in ALIGNNConv.forward(self, g, lg, x, y, z)
168 x, m = self.node_update(g, x, y)
170 # Edge-gated graph convolution update on crystal graph
--> 171 y, z = self.edge_update(lg, m, z)
173 return x, y, z
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/torch/nn/modules/module.py:1130, in Module._call_impl(self, *input, **kwargs)
1126 # If we don't have any hooks, we want to skip the rest of the logic in
1127 # this function, and just call forward.
1128 if not (self._backward_hooks or self._forward_hooks or self._forward_pre_hooks or _global_backward_hooks
1129 or _global_forward_hooks or _global_forward_pre_hooks):
-> 1130 return forward_call(*input, **kwargs)
1131 # Do not call functions when jit is used
1132 full_backward_hooks, non_full_backward_hooks = [], []
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/alignn/models/alignn_atomwise.py:111, in EdgeGatedGraphConv.forward(self, g, node_feats, edge_feats)
109 g.edata["sigma"] = torch.sigmoid(m)
110 g.ndata["Bh"] = self.dst_update(node_feats)
--> 111 g.update_all(
112 fn.u_mul_e("Bh", "sigma", "m"), fn.sum("m", "sum_sigma_h")
113 )
114 g.update_all(fn.copy_e("sigma", "m"), fn.sum("m", "sum_sigma"))
115 g.ndata["h"] = g.ndata["sum_sigma_h"] / (g.ndata["sum_sigma"] + 1e-6)
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/dgl/heterograph.py:4895, in DGLHeteroGraph.update_all(self, message_func, reduce_func, apply_node_func, etype)
4893 _, dtid = self._graph.metagraph.find_edge(etid)
4894 g = self if etype is None else self[etype]
-> 4895 ndata = core.message_passing(g, message_func, reduce_func, apply_node_func)
4896 if core.is_builtin(reduce_func) and reduce_func.name in ['min', 'max'] and ndata:
4897 # Replace infinity with zero for isolated nodes
4898 key = list(ndata.keys())[0]
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/dgl/core.py:357, in message_passing(g, mfunc, rfunc, afunc)
336 """Invoke message passing computation on the whole graph.
337
338 Parameters
(...)
352 Results from the message passing computation.
353 """
354 if (is_builtin(mfunc) and is_builtin(rfunc) and
355 getattr(ops, '{}_{}'.format(mfunc.name, rfunc.name), None) is not None):
356 # invoke fused message passing
--> 357 ndata = invoke_gspmm(g, mfunc, rfunc)
358 else:
359 # invoke message passing in two separate steps
360 # message phase
361 if is_builtin(mfunc):
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/dgl/core.py:323, in invoke_gspmm(graph, mfunc, rfunc, srcdata, dstdata, edata)
321 x = data_dict_to_list(graph, x, mfunc, lhs_target)
322 y = data_dict_to_list(graph, y, mfunc, rhs_target)
--> 323 z = op(graph, x, y)
324 else:
325 x = alldata[mfunc.target][mfunc.in_field]
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/dgl/ops/spmm.py:147, in _gen_spmm_func.<locals>.func(g, x, y)
146 def func(g, x, y):
--> 147 return gspmm(g, binary_op, reduce_op, x, y)
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/dgl/ops/spmm.py:75, in gspmm(g, op, reduce_op, lhs_data, rhs_data)
73 lhs_data, rhs_data = reshape_lhs_rhs(lhs_data, rhs_data)
74 # With max and min reducers infinity will be returned for zero degree nodes
---> 75 ret = gspmm_internal(g._graph, op,
76 'sum' if reduce_op == 'mean' else reduce_op,
77 lhs_data, rhs_data)
78 else:
79 # lhs_data or rhs_data is None only in unary functions like ``copy-u`` or ``copy_e``
80 lhs_data = [None] * g._graph.number_of_ntypes() if lhs_data is None else lhs_data
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/dgl/backend/pytorch/sparse.py:724, in gspmm(gidx, op, reduce_op, lhs_data, rhs_data)
722 op = 'mul'
723 rhs_data = 1. / rhs_data
--> 724 return GSpMM.apply(gidx, op, reduce_op, lhs_data, rhs_data)
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/torch/cuda/amp/autocast_mode.py:118, in custom_fwd.<locals>.decorate_fwd(*args, **kwargs)
116 return fwd(*_cast(args, cast_inputs), **_cast(kwargs, cast_inputs))
117 else:
--> 118 return fwd(*args, **kwargs)
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/dgl/backend/pytorch/sparse.py:106, in GSpMM.forward(ctx, gidx, op, reduce_op, X, Y)
103 @staticmethod
104 @custom_fwd(cast_inputs=th.float16)
105 def forward(ctx, gidx, op, reduce_op, X, Y):
--> 106 out, (argX, argY) = _gspmm(gidx, op, reduce_op, X, Y)
107 reduce_last = _need_reduce_last_dim(X, Y)
108 X_shape = X.shape if X is not None else None
File /media/colin/Shared/colin/git/materials-ml/env/lib/python3.10/site-packages/dgl/sparse.py:228, in _gspmm(gidx, op, reduce_op, u, e)
226 arg_e_nd = to_dgl_nd_for_write(arg_e)
227 if gidx.number_of_edges(0) > 0:
--> 228 _CAPI_DGLKernelSpMM(gidx, op, reduce_op,
229 to_dgl_nd(u if use_u else None),
230 to_dgl_nd(e if use_e else None),
231 to_dgl_nd_for_write(v),
232 arg_u_nd,
233 arg_e_nd)
234 # NOTE(zihao): actually we can avoid the following step, because arg_*_nd
235 # refers to the data that stores arg_*. After we call _CAPI_DGLKernelSpMM,
236 # arg_* should have already been changed. But we found this doesn't work
(...)
239 # The workaround is proposed by Jinjing, and we still need to investigate
240 # where the problem is.
241 arg_u = None if arg_u is None else F.zerocopy_from_dgl_ndarray(arg_u_nd)
KeyboardInterrupt: